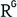Linkage disequilibrium score regression (LDSC) to quantify the extent of shared genetic contributions across 23 brain disorders (n=842,820), 11 quantitative and four dichotomous traits of interest (n=722,125) based on genome-wide association meta-analyses. Psychiatric disorders show substantial sharing of common variant risk, while many neurological disorders appear more distinct from one another,suggesting substantive differences in the specificity of the genetic etiology of these disorders. The high degree of genetic correlation among the psychiatric traits suggests that genetic risk factors for psychiatric disorders do not respect clinical diagnostic boundaries, congruent with the clinical controversies in classification with the clinical controversies in classification. The broad and continuous nature of psychiatric disorder spectra have been clinically recognized for a long time and these results suggest that shared biological mechanisms substantially contribute across psychiatric diagnoses.
O’Donovan, M., & Owen, M. (2016). The implications of the shared genetics of psychiatric disorders. Nature medicine, 22(11), 1214–1219.
Recent genomic studies have revealed the highly polygenic nature of psychiatric disorders, including schizophrenia, bipolar disorder and major depressive disorder. Many of the individual genetic associations are shared across multiple disorders in a way that points to extensive biological pleiotropy and further challenges the biological validity of existing diagnostic approaches. Here we argue that the existence of risk alleles specific to a single diagnostic category is unlikely. We also highlight some of the important clinical repercussions of pleiotropy.
Genetic correlation between schizophrenia and selected psychiatric disorders. Psychiatric disorders showing significant evidence (P ≤ 0.001) for overlaps between common variant contributions to schizophrenia and other psychiatric disorders. Overlaps are expressed as correlation in heritability (rg) captured by SNPs. Data are from ref. (39). ADHD, attention-deficit hyperactivity disorder; OCD, obsessive-compulsive disorder.
39. Antilla, V. et al. Analysis of shared heritability in common disorders of the brain. Preprint at bioRxiv http://dx.doi.org/10.1101/048991 (2016).
Drysdale, A., Grosenick, L., Downar, J., Dunlop, K., Mansouri, F., Meng, Y., Fetcho, R., et al. (2016). Resting-state connectivity biomarkers define neurophysiological subtypes of depression. Nature Medicine, 23(1), 28–38
Biotypes and biomarkers for depression.
(a) In a discovery sample, Drysdale et al. identified fMRI-imaging depression biotypes on the basis of patterns of functional connectivity, first by using canonical correlation analysis (CCA) to relate patterns of brain connectivity with symptom profiles, and then by clustering these individuals according to these connectivity patterns or components.
The biotypes were further optimized and accuracy was tested through cross-validated analysis (n = 711) and in an independent replication sample (n = 477). (b) Biotypes predicted response to dorsomedial prefrontal cortex (dmPFC) transcranial magnetic stimulation (TMS) treatment for 154 individuals with depression. (c) Biotypes of depression (dep.) overlapped with generalized anxiety disorder (GAD) but not schizophrenia (Sz.).
Drysdale, A., Grosenick, L., Downar, J., Dunlop, K., Mansouri, F., Meng, Y., Fetcho, R., et al. (2016). Resting-state connectivity biomarkers define neurophysiological subtypes of depression. Nature Medicine, 23(1), 28–38
Biotype-specific clinical profiles for the six depressive symptoms that varied most significantly by cluster (P < 0.005, Kruskal–Wallis ANOVA). Symptom severities (HAMD) are z-scored with respect to the mean for all patients in the cluster-discovery set.. (g) Boxplot of biotype differences in overall depression severity (total HAMD score), in which boxes denote the median and interquartile range (IQR) and whiskers the minimum and maximum values. In f and g, asterisk (*) indicates significant difference from mean symptom severity rating for all patients (z = 0) at P < 0.05; error bars depict s.e.m.; n.s., not significant.
Depression biotypes transcend conventional diagnostic boundaries
GAD was associated with widespread connectivity differences in resting-state networks (Fig. 5a–c) that overlapped significantly with those in depression (χ2 = 5,457; P < 0.0001; Fig. 5a–c)
Summary
Although major depressive depression—is up to 45% heritable (54), identifying genetic risk factors has proven challenging, even in extremely large genome-wide association studies (55). Likewise, efforts to develop new treatments have slowed, owing in part to a lack of physiological targets for the assessment of treatment efficacy and the selection of individuals who are most likely to benefit (56).
(54) Sullivan, P.F., Neale, M.C. & Kendler, K.S. Genetic epidemiology of major depression: review and meta-analysis. Am. J. Psychiatry 157, 1552–1562 (2000).
(55) Ripke, S. et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol. Psychiatry 18, 497–511 (2013).
(56) Pankevich, D.E., Altevogt, B.M., Dunlop, J., Gage, F.H. & Hyman, S.E. Improving and accelerating drug development for nervous system disorders. Neuron 84, 546–553 (2014).
Carving nature at the joints: sub- typing depression